Tabular analysis#
Thus far, we’ve read the data files as output by the National Water Model, which are provided as hourly snapshots in NetCDF format (or slight transformations of them, like via Kerchunk index files or conversion to Zarr).
A subset of the data (we’ll look at reservoir data) are distributed as NetCDF files but are fundamentally tabular. As an exercise, I converted some reservoir data to a geoparquet dataset. Parquet is a nice, high-performance dataset for tabular data. Geoparquet is like regular parquet, only with some extra geospatial metadata.
import dask.dataframe as dd
import dask_geopandas
import geopandas
This is a pretty large dataset, partitioned across many individual parquet files, so we’ll load it up with dask.dataframe / dask-geopandas.
df = dd.read_parquet(
"az://ciroh/short-range-reservoir.parquet/",
storage_options={"account_name": "noaanwm"},
calculate_divisions=True,
)
geometry = df.geometry.map_partitions(
geopandas.GeoSeries.from_wkb,
meta=geopandas.GeoSeries([], name="geometry"),
crs="epsg:4326",
)
gdf = dask_geopandas.from_dask_dataframe(df, geometry=geometry)
gdf
| feature_id | geometry | reservoir_type | water_sfc_elev | inflow | outflow | |
|---|---|---|---|---|---|---|
| npartitions=21 | ||||||
| 2021-08-26 01:00:00 | int32 | geometry | category[unknown] | float32 | float64 | float64 |
| 2021-09-01 01:00:00 | ... | ... | ... | ... | ... | ... |
| ... | ... | ... | ... | ... | ... | ... |
| 2023-04-01 01:00:00 | ... | ... | ... | ... | ... | ... |
| 2023-04-18 23:00:00 | ... | ... | ... | ... | ... | ... |
This data structure shows our 6 columns and their data types.
The output also shows that we have 21 partitions (or individual parquet files). I partitioned the data by month, so we can quickly look up a subset of the data.
w = gdf.loc["2022-07-01":"2022-07-08"]
w.head()
| feature_id | geometry | reservoir_type | water_sfc_elev | inflow | outflow | |
|---|---|---|---|---|---|---|
| time | ||||||
| 2022-07-01 | 491 | POINT (-68.37904 46.18327) | 1 | 206.158081 | 0.33 | 0.55 |
| 2022-07-01 | 531 | POINT (-68.45489 46.16116) | 1 | 247.600632 | 0.03 | 0.31 |
| 2022-07-01 | 747 | POINT (-68.06499 46.03409) | 1 | 190.322128 | 0.02 | 0.10 |
| 2022-07-01 | 759 | POINT (-68.16213 46.02238) | 1 | 165.124863 | 0.00 | 0.17 |
| 2022-07-01 | 1581 | POINT (-67.93720 45.64844) | 1 | 130.030273 | 0.48 | 0.55 |
We can do dataframe-style computations, like grouping by feature ID and getting some summary statistics for the inflow variable.
%%time
inflow = (
df.groupby("feature_id")
.inflow.agg(["mean", "min", "max", "std", "count"])
.compute()
)
inflow.head()
CPU times: user 7.45 s, sys: 2.05 s, total: 9.51 s
Wall time: 3.77 s
| mean | min | max | std | count | |
|---|---|---|---|---|---|
| feature_id | |||||
| 491 | 0.794296 | 0.07 | 15.20 | 1.006294 | 13236 |
| 531 | 0.342121 | 0.00 | 18.43 | 0.746961 | 13236 |
| 747 | 0.416411 | 0.01 | 14.90 | 0.815884 | 13236 |
| 759 | 0.000389 | 0.00 | 0.13 | 0.004175 | 13236 |
| 1581 | 1.271340 | 0.37 | 5.22 | 1.113017 | 13236 |
Let’s find the feature_id with the largest change in water elevation in the first month.
xs = gdf.get_partition(0).compute()
change = (
xs.groupby("feature_id")
.water_sfc_elev.agg(["min", "max"])
.diff(axis="columns")["max"]
)
feature_id = change.idxmax()
feature_id
22472505
We can get the geometry for that feature ID.
from ipyleaflet import Map, CircleMarker
geometry = xs[xs.feature_id == feature_id].iloc[0].geometry
y, x = geometry.y, geometry.x
m = Map(center=(y, x), zoom=15)
m.add(CircleMarker(location=(y, x)))
m.scroll_wheel_zoom = True
m
Now let’s examine the timeseries for that particular feature_id.
With dask.dataframe.read_parquet and pyarrow, we can supply a filters argument to efficiently filter the data based on the values, to avoid loading data we don’t care about.
ts = df = dd.read_parquet(
"az://ciroh/short-range-reservoir.parquet/",
storage_options={"account_name": "noaanwm"},
calculate_divisions=True,
filters=[("feature_id", "=", feature_id)],
columns=["inflow", "outflow", "water_sfc_elev"],
)
ts
| inflow | outflow | water_sfc_elev | |
|---|---|---|---|
| npartitions=21 | |||
| 2021-08-26 01:00:00 | float64 | float64 | float32 |
| 2021-09-01 01:00:00 | ... | ... | ... |
| ... | ... | ... | ... |
| 2023-04-01 01:00:00 | ... | ... | ... |
| 2023-04-18 23:00:00 | ... | ... | ... |
%time ts = ts.compute()
CPU times: user 5.17 s, sys: 1.82 s, total: 6.98 s
Wall time: 4.13 s
len(ts)
13236
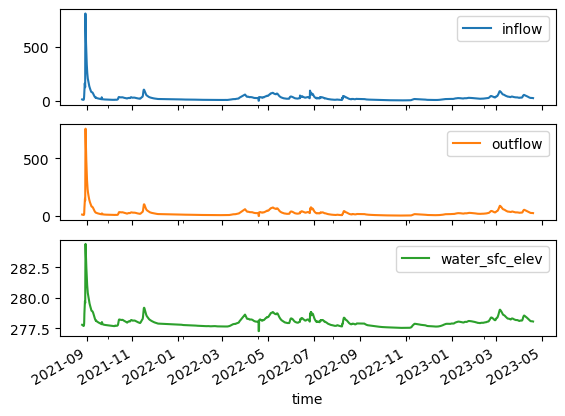
ts.plot(subplots=True);
Exercise: Explore more#
Spend some more time exploring this dataset. The next two cells get the locations where these measurements are taken.
Consider fusing this data with precipitation data from the noaa-mrms-qpe collection, or other products from the National Water Model.
points = xs.groupby("feature_id").geometry.first().set_crs("epsg:4326")
points.head()
feature_id
491 POINT (-68.37904 46.18327)
531 POINT (-68.45489 46.16116)
747 POINT (-68.06499 46.03409)
759 POINT (-68.16213 46.02238)
1581 POINT (-67.93720 45.64844)
Name: geometry, dtype: geometry
# Limit to 1,000 to avoid plotting too many points.
# You can filter spatially with .cx, or look at other subsets
points.head(1000).reset_index().explore()
