Kerchunk Background
In the last notebook, we saw that accessing data from the NetCDF file over the network was slow, in part because it was making a bunch of HTTP requests just to read some metadata that’s scattered around the NetCDF file. With a Kerchunk index file, you get to bypass all that seeking around for metadata: it’s already been extracted into the index file. While that’s maybe not a huge deal for a single NetCDF file, it matters a bunch when you’re dealing with thousands of NetCDF files (1,000 files * 1.5 seconds per file = ~25 minutes just to read metadata ).
<xarray.Dataset>
Dimensions: (time: 6866, feature_id: 2776738, reference_time: 1)
Coordinates:
* feature_id (feature_id) float64 101.0 179.0 181.0 ... 1.18e+09 1.18e+09
* reference_time (reference_time) datetime64[ns] 2022-06-29
* time (time) datetime64[ns] 2022-06-29T01:00:00 ... 2023-04-21T...
Data variables:
crs (time) object dask.array<chunksize=(1,), meta=np.ndarray>
nudge (time, feature_id) float64 dask.array<chunksize=(1, 2776738), meta=np.ndarray>
qBtmVertRunoff (time, feature_id) float64 dask.array<chunksize=(1, 2776738), meta=np.ndarray>
qBucket (time, feature_id) float64 dask.array<chunksize=(1, 2776738), meta=np.ndarray>
qSfcLatRunoff (time, feature_id) float64 dask.array<chunksize=(1, 2776738), meta=np.ndarray>
streamflow (time, feature_id) float64 dask.array<chunksize=(1, 2776738), meta=np.ndarray>
velocity (time, feature_id) float64 dask.array<chunksize=(1, 2776738), meta=np.ndarray>
Attributes: (12/19)
Conventions: CF-1.6
NWM_version_number: v2.2
TITLE: OUTPUT FROM NWM v2.2
cdm_datatype: Station
code_version: v5.2.0-beta2
dev: dev_ prefix indicates development/internal me...
... ...
model_output_type: channel_rt
model_output_valid_time: 2022-06-29_01:00:00
model_total_valid_times: 18
proj4: +proj=lcc +units=m +a=6370000.0 +b=6370000.0 ...
station_dimension: feature_id
stream_order_output: 1 Dimensions: time : 6866feature_id : 2776738reference_time : 1
Coordinates: (3)
Data variables: (7)
crs
(time)
object
dask.array<chunksize=(1,), meta=np.ndarray>
_CoordinateAxes : latitude longitude esri_pe_string : GEOGCS["GCS_WGS_1984",DATUM["D_WGS_1984",SPHEROID["WGS_1984",6378137.0,298.257223563]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]];-400 -400 1000000000;-100000 10000;-100000 10000;8.98315284119521E-09;0.001;0.001;IsHighPrecision grid_mapping_name : latitude longitude inverse_flattening : 298.2572326660156 long_name : CRS definition longitude_of_prime_meridian : 0.0 semi_major_axis : 6378137.0 semi_minor_axis : 6356752.5 spatial_ref : GEOGCS["GCS_WGS_1984",DATUM["D_WGS_1984",SPHEROID["WGS_1984",6378137.0,298.257223563]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]];-400 -400 1000000000;-100000 10000;-100000 10000;8.98315284119521E-09;0.001;0.001;IsHighPrecision transform_name : latitude longitude
Array
Chunk
Bytes
53.64 kiB
8 B
Shape
(6866,)
(1,)
Dask graph
6866 chunks in 2 graph layers
Data type
object numpy.ndarray
6866
1
nudge
(time, feature_id)
float64
dask.array<chunksize=(1, 2776738), meta=np.ndarray>
coordinates : latitude longitude grid_mapping : crs long_name : Amount of stream flow alteration units : m3 s-1 valid_range : [-5000000, 5000000]
Array
Chunk
Bytes
142.05 GiB
21.18 MiB
Shape
(6866, 2776738)
(1, 2776738)
Dask graph
6866 chunks in 2 graph layers
Data type
float64 numpy.ndarray
2776738
6866
qBtmVertRunoff
(time, feature_id)
float64
dask.array<chunksize=(1, 2776738), meta=np.ndarray>
coordinates : latitude longitude grid_mapping : crs long_name : Runoff from bottom of soil to bucket units : m3 valid_range : [0, 20000000]
Array
Chunk
Bytes
142.05 GiB
21.18 MiB
Shape
(6866, 2776738)
(1, 2776738)
Dask graph
6866 chunks in 2 graph layers
Data type
float64 numpy.ndarray
2776738
6866
qBucket
(time, feature_id)
float64
dask.array<chunksize=(1, 2776738), meta=np.ndarray>
coordinates : latitude longitude grid_mapping : crs long_name : Flux from gw bucket units : m3 s-1 valid_range : [0, 2000000000]
Array
Chunk
Bytes
142.05 GiB
21.18 MiB
Shape
(6866, 2776738)
(1, 2776738)
Dask graph
6866 chunks in 2 graph layers
Data type
float64 numpy.ndarray
2776738
6866
qSfcLatRunoff
(time, feature_id)
float64
dask.array<chunksize=(1, 2776738), meta=np.ndarray>
coordinates : latitude longitude grid_mapping : crs long_name : Runoff from terrain routing units : m3 s-1 valid_range : [0, 2000000000]
Array
Chunk
Bytes
142.05 GiB
21.18 MiB
Shape
(6866, 2776738)
(1, 2776738)
Dask graph
6866 chunks in 2 graph layers
Data type
float64 numpy.ndarray
2776738
6866
streamflow
(time, feature_id)
float64
dask.array<chunksize=(1, 2776738), meta=np.ndarray>
coordinates : latitude longitude grid_mapping : crs long_name : River Flow units : m3 s-1 valid_range : [0, 5000000]
Array
Chunk
Bytes
142.05 GiB
21.18 MiB
Shape
(6866, 2776738)
(1, 2776738)
Dask graph
6866 chunks in 2 graph layers
Data type
float64 numpy.ndarray
2776738
6866
velocity
(time, feature_id)
float64
dask.array<chunksize=(1, 2776738), meta=np.ndarray>
coordinates : latitude longitude grid_mapping : crs long_name : River Velocity units : m s-1 valid_range : [0, 5000000]
Array
Chunk
Bytes
142.05 GiB
21.18 MiB
Shape
(6866, 2776738)
(1, 2776738)
Dask graph
6866 chunks in 2 graph layers
Data type
float64 numpy.ndarray
2776738
6866
Indexes: (3)
PandasIndex
PandasIndex(Index([ 101.0, 179.0, 181.0, 183.0, 185.0,
843.0, 845.0, 847.0, 849.0, 851.0,
...
1180001795.0, 1180001796.0, 1180001797.0, 1180001798.0, 1180001799.0,
1180001800.0, 1180001801.0, 1180001802.0, 1180001803.0, 1180001804.0],
dtype='float64', name='feature_id', length=2776738)) PandasIndex
PandasIndex(DatetimeIndex(['2022-06-29'], dtype='datetime64[ns]', name='reference_time', freq=None)) PandasIndex
PandasIndex(DatetimeIndex(['2022-06-29 01:00:00', '2022-06-29 02:00:00',
'2022-06-29 03:00:00', '2022-06-29 04:00:00',
'2022-06-29 05:00:00', '2022-06-29 06:00:00',
'2022-06-29 07:00:00', '2022-06-29 08:00:00',
'2022-06-29 09:00:00', '2022-06-29 10:00:00',
...
'2023-04-21 07:00:00', '2023-04-21 08:00:00',
'2023-04-21 09:00:00', '2023-04-21 10:00:00',
'2023-04-21 11:00:00', '2023-04-21 12:00:00',
'2023-04-21 13:00:00', '2023-04-21 14:00:00',
'2023-04-21 15:00:00', '2023-04-21 16:00:00'],
dtype='datetime64[ns]', name='time', length=6866, freq=None)) Attributes: (19)
Conventions : CF-1.6 NWM_version_number : v2.2 TITLE : OUTPUT FROM NWM v2.2 cdm_datatype : Station code_version : v5.2.0-beta2 dev : dev_ prefix indicates development/internal meta data dev_NOAH_TIMESTEP : 3600 dev_OVRTSWCRT : 1 dev_channelBucket_only : 0 dev_channel_only : 0 featureType : timeSeries model_configuration : short_range model_initialization_time : 2022-06-29_00:00:00 model_output_type : channel_rt model_output_valid_time : 2022-06-29_01:00:00 model_total_valid_times : 18 proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@ station_dimension : feature_id stream_order_output : 1
So in about a second, we opened up the Kerchunk index file and used that to build our Dataset. Doing that from the raw NetCDF files would have taken ~hours.
You’ll notice we’re using the zarr m
Zarr uses a simple dictionary-like interface with well-known keys. You can get the global attributes at .zattrs
Or the attributes for a specific array:
Those attributes saved in the Kerchunk index file, and so don’t require any (additional) HTTP requests to get. But we don’t want to re-save the large data variables, since we don’t want to host the data twice. So what happens when you want to read a data variable? Well, we can look at the references
In a typical Zarr dataset, accessing an array at <name>/0.0
The URL where this chunk of data comes from
The offset within that file
The size of the chunk of data, in bytes, on disk
Thanks to HTTP range requests (the same thing that powers streaming video) we can request just the subset of the file we need. When a high-level library like xarray asks the data in that chunk, this toolchain (of zarr, fsspec’s reference filesystem, and adlfs) will make the HTTP range request in the background and deliver the bytes.
One very important caveat: because Kerchunk is just an index on the existing data, we inherit all of the limitations of its chunking structure. This datsaet is chunked by hour along time
If you’re interested in generating the Kerchunk files, see the processing directory . I used the pangeo-forge library to write a “recipe” for generating Kerchunk index files from a bunch of NetCDF files, and stick them together properly. I then executed that job using the same Kubernetes cluster we’re using for this workshop.
Exercise: Have fun!
I’ve also created Kerchunk indexes for the 0-hour forecasts for land forcing
<xarray.Dataset>
Dimensions: (time: 6992, y: 3840, x: 4608, reference_time: 1)
Coordinates:
* reference_time (reference_time) datetime64[ns] 2022-06-29
* time (time) datetime64[ns] 2022-06-29T01:00:00 ... 2023-04-26T...
* x (x) float64 -2.303e+06 -2.302e+06 ... 2.303e+06 2.304e+06
* y (y) float64 -1.92e+06 -1.919e+06 ... 1.918e+06 1.919e+06
Data variables:
ACCET (time, y, x) float64 dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
FSNO (time, y, x) float64 dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
SNEQV (time, y, x) float64 dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
SNOWH (time, y, x) float64 dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
SNOWT_AVG (time, y, x) float64 dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
SOILSAT_TOP (time, y, x) float64 dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
crs (time) object dask.array<chunksize=(1,), meta=np.ndarray>
Attributes:
Conventions: CF-1.6
GDAL_DataType: Generic
NWM_version_number: v2.2
TITLE: OUTPUT FROM NWM v2.2
code_version: v5.2.0-beta2
model_configuration: short_range
model_initialization_time: 2022-06-29_00:00:00
model_output_type: land
model_output_valid_time: 2022-06-29_01:00:00
model_total_valid_times: 18
proj4: +proj=lcc +units=m +a=6370000.0 +b=6370000.0 ... Dimensions: time : 6992y : 3840x : 4608reference_time : 1
Coordinates: (4)
reference_time
(reference_time)
datetime64[ns]
2022-06-29
long_name : model initialization time standard_name : forecast_reference_time array(['2022-06-29T00:00:00.000000000'], dtype='datetime64[ns]') time
(time)
datetime64[ns]
2022-06-29T01:00:00 ... 2023-04-...
long_name : valid output time standard_name : time valid_max : 27608760 valid_min : 27607740 array(['2022-06-29T01:00:00.000000000', '2022-06-29T02:00:00.000000000',
'2022-06-29T03:00:00.000000000', ..., '2023-04-26T17:00:00.000000000',
'2023-04-26T18:00:00.000000000', '2023-04-26T19:00:00.000000000'],
dtype='datetime64[ns]') x
(x)
float64
-2.303e+06 -2.302e+06 ... 2.304e+06
_CoordinateAxisType : GeoX long_name : x coordinate of projection resolution : 1000.0 standard_name : projection_x_coordinate units : m array([-2303499.25, -2302499.25, -2301499.25, ..., 2301500.75, 2302500.75,
2303500.75]) y
(y)
float64
-1.92e+06 -1.919e+06 ... 1.919e+06
_CoordinateAxisType : GeoY long_name : y coordinate of projection resolution : 1000.0 standard_name : projection_y_coordinate units : m array([-1919500.375, -1918500.375, -1917500.375, ..., 1917499.625,
1918499.625, 1919499.625]) Data variables: (7)
ACCET
(time, y, x)
float64
dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Accumulated total ET units : mm valid_range : [-100000, 100000000]
Array
Chunk
Bytes
0.90 TiB
5.40 MiB
Shape
(6992, 3840, 4608)
(1, 768, 922)
Dask graph
174800 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
6992
FSNO
(time, y, x)
float64
dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Snow-cover fraction on the ground units : 1 valid_range : [0, 1000]
Array
Chunk
Bytes
0.90 TiB
5.40 MiB
Shape
(6992, 3840, 4608)
(1, 768, 922)
Dask graph
174800 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
6992
SNEQV
(time, y, x)
float64
dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Snow water equivalent units : kg m-2 valid_range : [0, 1000000]
Array
Chunk
Bytes
0.90 TiB
5.40 MiB
Shape
(6992, 3840, 4608)
(1, 768, 922)
Dask graph
174800 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
6992
SNOWH
(time, y, x)
float64
dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Snow depth units : m valid_range : [0, 1000000]
Array
Chunk
Bytes
0.90 TiB
5.40 MiB
Shape
(6992, 3840, 4608)
(1, 768, 922)
Dask graph
174800 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
6992
SNOWT_AVG
(time, y, x)
float64
dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : average snow temperature (by layer mass) units : K valid_range : [0, 4000]
Array
Chunk
Bytes
0.90 TiB
5.40 MiB
Shape
(6992, 3840, 4608)
(1, 768, 922)
Dask graph
174800 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
6992
SOILSAT_TOP
(time, y, x)
float64
dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : fraction of soil saturation, top 2 layers units : 1 valid_range : [0, 1000]
Array
Chunk
Bytes
0.90 TiB
5.40 MiB
Shape
(6992, 3840, 4608)
(1, 768, 922)
Dask graph
174800 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
6992
crs
(time)
object
dask.array<chunksize=(1,), meta=np.ndarray>
GeoTransform : -2303999.17655 1000.0 0 1919999.66329 0 -1000.0 _CoordinateAxes : y x _CoordinateTransformType : Projection earth_radius : 6370000.0 esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision false_easting : 0.0 false_northing : 0.0 grid_mapping_name : lambert_conformal_conic inverse_flattening : 0.0 latitude_of_projection_origin : 40.0 long_name : CRS definition longitude_of_central_meridian : -97.0 longitude_of_prime_meridian : 0.0 semi_major_axis : 6370000.0 spatial_ref : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision standard_parallel : [30.0, 60.0] transform_name : lambert_conformal_conic
Array
Chunk
Bytes
54.62 kiB
8 B
Shape
(6992,)
(1,)
Dask graph
6992 chunks in 2 graph layers
Data type
object numpy.ndarray
6992
1
Indexes: (4)
PandasIndex
PandasIndex(DatetimeIndex(['2022-06-29'], dtype='datetime64[ns]', name='reference_time', freq=None)) PandasIndex
PandasIndex(DatetimeIndex(['2022-06-29 01:00:00', '2022-06-29 02:00:00',
'2022-06-29 03:00:00', '2022-06-29 04:00:00',
'2022-06-29 05:00:00', '2022-06-29 06:00:00',
'2022-06-29 07:00:00', '2022-06-29 08:00:00',
'2022-06-29 09:00:00', '2022-06-29 10:00:00',
...
'2023-04-26 10:00:00', '2023-04-26 11:00:00',
'2023-04-26 12:00:00', '2023-04-26 13:00:00',
'2023-04-26 14:00:00', '2023-04-26 15:00:00',
'2023-04-26 16:00:00', '2023-04-26 17:00:00',
'2023-04-26 18:00:00', '2023-04-26 19:00:00'],
dtype='datetime64[ns]', name='time', length=6992, freq=None)) PandasIndex
PandasIndex(Index([-2303499.25, -2302499.25, -2301499.25, -2300499.25, -2299499.25,
-2298499.25, -2297499.25, -2296499.25, -2295499.25, -2294499.25,
...
2294500.75, 2295500.75, 2296500.75, 2297500.75, 2298500.75,
2299500.75, 2300500.75, 2301500.75, 2302500.75, 2303500.75],
dtype='float64', name='x', length=4608)) PandasIndex
PandasIndex(Index([-1919500.375, -1918500.375, -1917500.375, -1916500.375, -1915500.375,
-1914500.375, -1913500.375, -1912500.375, -1911500.375, -1910500.375,
...
1910499.625, 1911499.625, 1912499.625, 1913499.625, 1914499.625,
1915499.625, 1916499.625, 1917499.625, 1918499.625, 1919499.625],
dtype='float64', name='y', length=3840)) Attributes: (11)
Conventions : CF-1.6 GDAL_DataType : Generic NWM_version_number : v2.2 TITLE : OUTPUT FROM NWM v2.2 code_version : v5.2.0-beta2 model_configuration : short_range model_initialization_time : 2022-06-29_00:00:00 model_output_type : land model_output_valid_time : 2022-06-29_01:00:00 model_total_valid_times : 18 proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs
<xarray.Dataset>
Dimensions: (time: 7306, y: 3840, x: 4608, reference_time: 1)
Coordinates:
* reference_time (reference_time) datetime64[ns] 2022-06-29
* time (time) datetime64[ns] 2022-06-29T01:00:00 ... 2023-04-29T...
* x (x) float64 -2.303e+06 -2.302e+06 ... 2.303e+06 2.304e+06
* y (y) float64 -1.92e+06 -1.919e+06 ... 1.918e+06 1.919e+06
Data variables:
LWDOWN (time, y, x) float64 dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
PSFC (time, y, x) float64 dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
Q2D (time, y, x) float64 dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
RAINRATE (time, y, x) float32 dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
SWDOWN (time, y, x) float64 dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
T2D (time, y, x) float64 dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
U2D (time, y, x) float64 dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
V2D (time, y, x) float64 dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
crs object ...
Attributes:
NWM_version_number: v2.2
model_configuration: short_range
model_initialization_time: 2022-06-29_00:00:00
model_output_type: forcing
model_output_valid_time: 2022-06-29_01:00:00
model_total_valid_times: 18 Dimensions: time : 7306y : 3840x : 4608reference_time : 1
Coordinates: (4)
reference_time
(reference_time)
datetime64[ns]
2022-06-29
long_name : model initialization time standard_name : forecast_reference_time array(['2022-06-29T00:00:00.000000000'], dtype='datetime64[ns]') time
(time)
datetime64[ns]
2022-06-29T01:00:00 ... 2023-04-...
long_name : valid output time standard_name : time array(['2022-06-29T01:00:00.000000000', '2022-06-29T02:00:00.000000000',
'2022-06-29T03:00:00.000000000', ..., '2023-04-29T09:00:00.000000000',
'2023-04-29T10:00:00.000000000', '2023-04-29T11:00:00.000000000'],
dtype='datetime64[ns]') x
(x)
float64
-2.303e+06 -2.302e+06 ... 2.304e+06
_CoordinateAxisType : GeoX long_name : x coordinate of projection resolution : 1000.0 standard_name : projection_x_coordinate units : m array([-2303499.25, -2302499.25, -2301499.25, ..., 2301500.75, 2302500.75,
2303500.75]) y
(y)
float64
-1.92e+06 -1.919e+06 ... 1.919e+06
_CoordinateAxisType : GeoY long_name : y coordinate of projection resolution : 1000.0 standard_name : projection_y_coordinate units : m array([-1919500.375, -1918500.375, -1917500.375, ..., 1917499.625,
1918499.625, 1919499.625]) Data variables: (9)
LWDOWN
(time, y, x)
float64
dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Surface downward long-wave radiation flux proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : surface_downward_longwave_flux units : W m-2
Array
Chunk
Bytes
0.94 TiB
5.40 MiB
Shape
(7306, 3840, 4608)
(1, 768, 922)
Dask graph
182650 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
7306
PSFC
(time, y, x)
float64
dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Surface Pressure proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : air_pressure units : Pa
Array
Chunk
Bytes
0.94 TiB
5.40 MiB
Shape
(7306, 3840, 4608)
(1, 768, 922)
Dask graph
182650 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
7306
Q2D
(time, y, x)
float64
dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : 2-m Specific Humidity proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : surface_specific_humidity units : kg kg-1
Array
Chunk
Bytes
0.94 TiB
5.40 MiB
Shape
(7306, 3840, 4608)
(1, 768, 922)
Dask graph
182650 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
7306
RAINRATE
(time, y, x)
float32
dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
cell_methods : time: mean esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Surface Precipitation Rate proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : precipitation_flux units : mm s^-1
Array
Chunk
Bytes
481.60 GiB
2.70 MiB
Shape
(7306, 3840, 4608)
(1, 768, 922)
Dask graph
182650 chunks in 2 graph layers
Data type
float32 numpy.ndarray
4608
3840
7306
SWDOWN
(time, y, x)
float64
dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
cell_methods : time point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Surface downward short-wave radiation flux proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : surface_downward_shortwave_flux units : W m-2
Array
Chunk
Bytes
0.94 TiB
5.40 MiB
Shape
(7306, 3840, 4608)
(1, 768, 922)
Dask graph
182650 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
7306
T2D
(time, y, x)
float64
dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : 2-m Air Temperature proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : air_temperature units : K
Array
Chunk
Bytes
0.94 TiB
5.40 MiB
Shape
(7306, 3840, 4608)
(1, 768, 922)
Dask graph
182650 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
7306
U2D
(time, y, x)
float64
dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : 10-m U-component of wind proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : x_wind units : m s-1
Array
Chunk
Bytes
0.94 TiB
5.40 MiB
Shape
(7306, 3840, 4608)
(1, 768, 922)
Dask graph
182650 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
7306
V2D
(time, y, x)
float64
dask.array<chunksize=(1, 768, 922), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : 10-m V-component of wind proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : y_wind units : m s-1
Array
Chunk
Bytes
0.94 TiB
5.40 MiB
Shape
(7306, 3840, 4608)
(1, 768, 922)
Dask graph
182650 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
7306
crs
()
object
...
GeoTransform : -2303999.17655 1000.0 0 1919999.66329 0 -1000.0 _CoordinateAxes : y x _CoordinateTransformType : Projection earth_radius : 6370000.0 esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision false_easting : 0.0 false_northing : 0.0 grid_mapping_name : lambert_conformal_conic inverse_flattening : 0.0 latitude_of_projection_origin : 40.0 long_name : CRS definition longitude_of_central_meridian : -97.0 longitude_of_prime_meridian : 0.0 semi_major_axis : 6370000.0 spatial_ref : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision standard_parallel : [30.0, 60.0] transform_name : lambert_conformal_conic [1 values with dtype=object] Indexes: (4)
PandasIndex
PandasIndex(DatetimeIndex(['2022-06-29'], dtype='datetime64[ns]', name='reference_time', freq=None)) PandasIndex
PandasIndex(DatetimeIndex(['2022-06-29 01:00:00', '2022-06-29 02:00:00',
'2022-06-29 03:00:00', '2022-06-29 04:00:00',
'2022-06-29 05:00:00', '2022-06-29 06:00:00',
'2022-06-29 07:00:00', '2022-06-29 08:00:00',
'2022-06-29 09:00:00', '2022-06-29 10:00:00',
...
'2023-04-29 02:00:00', '2023-04-29 03:00:00',
'2023-04-29 04:00:00', '2023-04-29 05:00:00',
'2023-04-29 06:00:00', '2023-04-29 07:00:00',
'2023-04-29 08:00:00', '2023-04-29 09:00:00',
'2023-04-29 10:00:00', '2023-04-29 11:00:00'],
dtype='datetime64[ns]', name='time', length=7306, freq=None)) PandasIndex
PandasIndex(Index([-2303499.25, -2302499.25, -2301499.25, -2300499.25, -2299499.25,
-2298499.25, -2297499.25, -2296499.25, -2295499.25, -2294499.25,
...
2294500.75, 2295500.75, 2296500.75, 2297500.75, 2298500.75,
2299500.75, 2300500.75, 2301500.75, 2302500.75, 2303500.75],
dtype='float64', name='x', length=4608)) PandasIndex
PandasIndex(Index([-1919500.375, -1918500.375, -1917500.375, -1916500.375, -1915500.375,
-1914500.375, -1913500.375, -1912500.375, -1911500.375, -1910500.375,
...
1910499.625, 1911499.625, 1912499.625, 1913499.625, 1914499.625,
1915499.625, 1916499.625, 1917499.625, 1918499.625, 1919499.625],
dtype='float64', name='y', length=3840)) Attributes: (6)
NWM_version_number : v2.2 model_configuration : short_range model_initialization_time : 2022-06-29_00:00:00 model_output_type : forcing model_output_valid_time : 2022-06-29_01:00:00 model_total_valid_times : 18
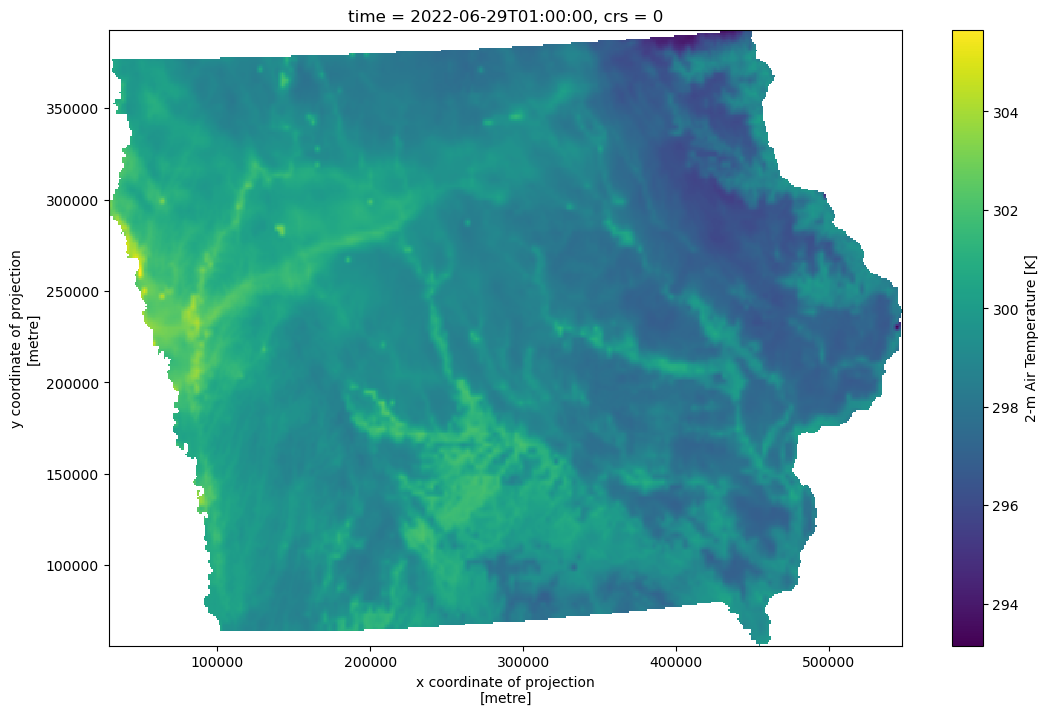
As an example, let’s load up the state boundary for Iowa from the Planetary Computer’s US Census dataset .
We can clip the full forcing
<xarray.DataArray 'T2D' (time: 7306, y: 337, x: 519)>
dask.array<getitem, shape=(7306, 337, 519), dtype=float64, chunksize=(1, 328, 433), chunktype=numpy.ndarray>
Coordinates:
* time (time) datetime64[ns] 2022-06-29T01:00:00 ... 2023-04-29T11:00:00
* x (x) float64 2.95e+04 3.05e+04 3.15e+04 ... 5.465e+05 5.475e+05
* y (y) float64 5.65e+04 5.75e+04 5.85e+04 ... 3.915e+05 3.925e+05
crs int64 0
Attributes:
cell_methods: time: point
esri_pe_string: PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DAT...
grid_mapping: crs
long_name: 2-m Air Temperature
proj4: +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0...
remap: remapped via ESMF regrid_with_weights: Bilinear
standard_name: air_temperature
units: K Coordinates: (4)
time
(time)
datetime64[ns]
2022-06-29T01:00:00 ... 2023-04-...
long_name : valid output time standard_name : time array(['2022-06-29T01:00:00.000000000', '2022-06-29T02:00:00.000000000',
'2022-06-29T03:00:00.000000000', ..., '2023-04-29T09:00:00.000000000',
'2023-04-29T10:00:00.000000000', '2023-04-29T11:00:00.000000000'],
dtype='datetime64[ns]') x
(x)
float64
2.95e+04 3.05e+04 ... 5.475e+05
_CoordinateAxisType : GeoX long_name : x coordinate of projection resolution : 1000.0 standard_name : projection_x_coordinate units : metre axis : X array([ 29500.824219, 30500.824219, 31500.824219, ..., 545500.8125 ,
546500.8125 , 547500.8125 ]) y
(y)
float64
5.65e+04 5.75e+04 ... 3.925e+05
_CoordinateAxisType : GeoY long_name : y coordinate of projection resolution : 1000.0 standard_name : projection_y_coordinate units : metre axis : Y array([ 56499.664062, 57499.664062, 58499.664062, ..., 390499.65625 ,
391499.65625 , 392499.65625 ]) crs
()
int64
0
crs_wkt : PROJCS["Lambert_Conformal_Conic",GEOGCS["Unknown datum based upon the Authalic Sphere",DATUM["Not_specified_based_on_Authalic_Sphere",SPHEROID["Sphere",6370000,0],AUTHORITY["EPSG","6035"]],PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0],PARAMETER["false_northing",0],PARAMETER["central_meridian",-97],PARAMETER["standard_parallel_1",30],PARAMETER["standard_parallel_2",60],PARAMETER["latitude_of_origin",40],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH]] semi_major_axis : 6370000.0 semi_minor_axis : 6370000.0 inverse_flattening : 0.0 reference_ellipsoid_name : Sphere longitude_of_prime_meridian : 0.0 prime_meridian_name : Greenwich geographic_crs_name : Unknown datum based upon the Authalic Sphere horizontal_datum_name : Not specified (based on Authalic Sphere) projected_crs_name : Lambert_Conformal_Conic grid_mapping_name : lambert_conformal_conic standard_parallel : (30.0, 60.0) latitude_of_projection_origin : 40.0 longitude_of_central_meridian : -97.0 false_easting : 0.0 false_northing : 0.0 spatial_ref : PROJCS["Lambert_Conformal_Conic",GEOGCS["Unknown datum based upon the Authalic Sphere",DATUM["Not_specified_based_on_Authalic_Sphere",SPHEROID["Sphere",6370000,0],AUTHORITY["EPSG","6035"]],PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0],PARAMETER["false_northing",0],PARAMETER["central_meridian",-97],PARAMETER["standard_parallel_1",30],PARAMETER["standard_parallel_2",60],PARAMETER["latitude_of_origin",40],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH]] GeoTransform : 29000.824230061535 999.9999773769305 0.0 55999.66407412574 0.0 999.9999767485119 Indexes: (3)
PandasIndex
PandasIndex(DatetimeIndex(['2022-06-29 01:00:00', '2022-06-29 02:00:00',
'2022-06-29 03:00:00', '2022-06-29 04:00:00',
'2022-06-29 05:00:00', '2022-06-29 06:00:00',
'2022-06-29 07:00:00', '2022-06-29 08:00:00',
'2022-06-29 09:00:00', '2022-06-29 10:00:00',
...
'2023-04-29 02:00:00', '2023-04-29 03:00:00',
'2023-04-29 04:00:00', '2023-04-29 05:00:00',
'2023-04-29 06:00:00', '2023-04-29 07:00:00',
'2023-04-29 08:00:00', '2023-04-29 09:00:00',
'2023-04-29 10:00:00', '2023-04-29 11:00:00'],
dtype='datetime64[ns]', name='time', length=7306, freq=None)) PandasIndex
PandasIndex(Index([29500.82421875, 30500.82421875, 31500.82421875, 32500.82421875,
33500.82421875, 34500.82421875, 35500.82421875, 36500.82421875,
37500.82421875, 38500.82421875,
...
538500.8125, 539500.8125, 540500.8125, 541500.8125,
542500.8125, 543500.8125, 544500.8125, 545500.8125,
546500.8125, 547500.8125],
dtype='float64', name='x', length=519)) PandasIndex
PandasIndex(Index([56499.6640625, 57499.6640625, 58499.6640625, 59499.6640625,
60499.6640625, 61499.6640625, 62499.6640625, 63499.6640625,
64499.6640625, 65499.6640625,
...
383499.65625, 384499.65625, 385499.65625, 386499.65625,
387499.65625, 388499.65625, 389499.65625, 390499.65625,
391499.65625, 392499.65625],
dtype='float64', name='y', length=337)) Attributes: (8)
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : 2-m Air Temperature proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : air_temperature units : K
Spend a bit of time playing around with these datasets.
When you’re done here, close down the kernel and move on to Timeseries