Timeseries analysis
This notebook analyzes the National Water Model timeseries. Our previous notebook used datasets that were chunked in time, which enables fast access to a temporal snapshot of the entire Continental United States. For this notebook, we’ll used a rechunked dataset that’s (primarily) chunked in space.
<xarray.Dataset>
Dimensions: (time: 7306, y: 3840, x: 4608, reference_time: 1)
Coordinates:
crs int64 0
* reference_time (reference_time) datetime64[ns] 2023-04-27T11:00:00
* time (time) datetime64[ns] 2022-06-29 ... 2023-04-29T10:00:00
* x (x) float64 -2.303e+06 -2.302e+06 ... 2.303e+06 2.304e+06
* y (y) float64 -1.92e+06 -1.919e+06 ... 1.918e+06 1.919e+06
Data variables:
LWDOWN (time, y, x) float64 dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
PSFC (time, y, x) float64 dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
Q2D (time, y, x) float64 dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
RAINRATE (time, y, x) float32 dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
SWDOWN (time, y, x) float64 dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
T2D (time, y, x) float64 dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
U2D (time, y, x) float64 dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
V2D (time, y, x) float64 dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
Attributes:
NWM_version_number: v2.2
model_configuration: short_range
model_initialization_time: 2023-04-27_11:00:00
model_output_type: forcing
model_output_valid_time: 2023-04-27_12:00:00
model_total_valid_times: 18
pangeo-forge:inputs_hash: 59784c1feed1b881dc936b070ac5a83cbc680599cb9b6...
pangeo-forge:recipe_hash: 25e9980cd34a6a0871883fc7375f224d34f403c944f95...
pangeo-forge:version: 0.9.4 Dimensions: time : 7306y : 3840x : 4608reference_time : 1
Coordinates: (5)
crs
()
int64
0
crs_wkt : PROJCS["Lambert_Conformal_Conic",GEOGCS["Unknown datum based upon the Authalic Sphere",DATUM["Not_specified_based_on_Authalic_Sphere",SPHEROID["Sphere",6370000,0],AUTHORITY["EPSG","6035"]],PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0],PARAMETER["false_northing",0],PARAMETER["central_meridian",-97],PARAMETER["standard_parallel_1",30],PARAMETER["standard_parallel_2",60],PARAMETER["latitude_of_origin",40],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH]] semi_major_axis : 6370000.0 semi_minor_axis : 6370000.0 inverse_flattening : 0.0 reference_ellipsoid_name : Sphere longitude_of_prime_meridian : 0.0 prime_meridian_name : Greenwich geographic_crs_name : Unknown datum based upon the Authalic Sphere horizontal_datum_name : Not specified (based on Authalic Sphere) projected_crs_name : Lambert_Conformal_Conic grid_mapping_name : lambert_conformal_conic standard_parallel : (30.0, 60.0) latitude_of_projection_origin : 40.0 longitude_of_central_meridian : -97.0 false_easting : 0.0 false_northing : 0.0 spatial_ref : PROJCS["Lambert_Conformal_Conic",GEOGCS["Unknown datum based upon the Authalic Sphere",DATUM["Not_specified_based_on_Authalic_Sphere",SPHEROID["Sphere",6370000,0],AUTHORITY["EPSG","6035"]],PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0],PARAMETER["false_northing",0],PARAMETER["central_meridian",-97],PARAMETER["standard_parallel_1",30],PARAMETER["standard_parallel_2",60],PARAMETER["latitude_of_origin",40],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH]] reference_time
(reference_time)
datetime64[ns]
2023-04-27T11:00:00
long_name : model initialization time standard_name : forecast_reference_time array(['2023-04-27T11:00:00.000000000'], dtype='datetime64[ns]') time
(time)
datetime64[ns]
2022-06-29 ... 2023-04-29T10:00:00
array(['2022-06-29T00:00:00.000000000', '2022-06-29T01:00:00.000000000',
'2022-06-29T02:00:00.000000000', ..., '2023-04-29T08:00:00.000000000',
'2023-04-29T09:00:00.000000000', '2023-04-29T10:00:00.000000000'],
dtype='datetime64[ns]') x
(x)
float64
-2.303e+06 -2.302e+06 ... 2.304e+06
_CoordinateAxisType : GeoX long_name : x coordinate of projection resolution : 1000.0 standard_name : projection_x_coordinate units : m array([-2303499.25, -2302499.25, -2301499.25, ..., 2301500.75, 2302500.75,
2303500.75]) y
(y)
float64
-1.92e+06 -1.919e+06 ... 1.919e+06
_CoordinateAxisType : GeoY long_name : y coordinate of projection resolution : 1000.0 standard_name : projection_y_coordinate units : m array([-1919500.375, -1918500.375, -1917500.375, ..., 1917499.625,
1918499.625, 1919499.625]) Data variables: (8)
LWDOWN
(time, y, x)
float64
dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Surface downward long-wave radiation flux proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : surface_downward_longwave_flux units : W m-2
Array
Chunk
Bytes
0.94 TiB
88.59 MiB
Shape
(7306, 3840, 4608)
(168, 240, 288)
Dask graph
11264 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
7306
PSFC
(time, y, x)
float64
dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Surface Pressure proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : air_pressure units : Pa
Array
Chunk
Bytes
0.94 TiB
88.59 MiB
Shape
(7306, 3840, 4608)
(168, 240, 288)
Dask graph
11264 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
7306
Q2D
(time, y, x)
float64
dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : 2-m Specific Humidity proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : surface_specific_humidity units : kg kg-1
Array
Chunk
Bytes
0.94 TiB
88.59 MiB
Shape
(7306, 3840, 4608)
(168, 240, 288)
Dask graph
11264 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
7306
RAINRATE
(time, y, x)
float32
dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
cell_methods : time: mean esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Surface Precipitation Rate proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : precipitation_flux units : mm s^-1
Array
Chunk
Bytes
481.60 GiB
44.30 MiB
Shape
(7306, 3840, 4608)
(168, 240, 288)
Dask graph
11264 chunks in 2 graph layers
Data type
float32 numpy.ndarray
4608
3840
7306
SWDOWN
(time, y, x)
float64
dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
cell_methods : time point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Surface downward short-wave radiation flux proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : surface_downward_shortwave_flux units : W m-2
Array
Chunk
Bytes
0.94 TiB
88.59 MiB
Shape
(7306, 3840, 4608)
(168, 240, 288)
Dask graph
11264 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
7306
T2D
(time, y, x)
float64
dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : 2-m Air Temperature proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : air_temperature units : K
Array
Chunk
Bytes
0.94 TiB
88.59 MiB
Shape
(7306, 3840, 4608)
(168, 240, 288)
Dask graph
11264 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
7306
U2D
(time, y, x)
float64
dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : 10-m U-component of wind proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : x_wind units : m s-1
Array
Chunk
Bytes
0.94 TiB
88.59 MiB
Shape
(7306, 3840, 4608)
(168, 240, 288)
Dask graph
11264 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
7306
V2D
(time, y, x)
float64
dask.array<chunksize=(168, 240, 288), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : 10-m V-component of wind proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : y_wind units : m s-1
Array
Chunk
Bytes
0.94 TiB
88.59 MiB
Shape
(7306, 3840, 4608)
(168, 240, 288)
Dask graph
11264 chunks in 2 graph layers
Data type
float64 numpy.ndarray
4608
3840
7306
Indexes: (4)
PandasIndex
PandasIndex(DatetimeIndex(['2023-04-27 11:00:00'], dtype='datetime64[ns]', name='reference_time', freq=None)) PandasIndex
PandasIndex(DatetimeIndex(['2022-06-29 00:00:00', '2022-06-29 01:00:00',
'2022-06-29 02:00:00', '2022-06-29 03:00:00',
'2022-06-29 04:00:00', '2022-06-29 05:00:00',
'2022-06-29 06:00:00', '2022-06-29 07:00:00',
'2022-06-29 08:00:00', '2022-06-29 09:00:00',
...
'2023-04-29 01:00:00', '2023-04-29 02:00:00',
'2023-04-29 03:00:00', '2023-04-29 04:00:00',
'2023-04-29 05:00:00', '2023-04-29 06:00:00',
'2023-04-29 07:00:00', '2023-04-29 08:00:00',
'2023-04-29 09:00:00', '2023-04-29 10:00:00'],
dtype='datetime64[ns]', name='time', length=7306, freq=None)) PandasIndex
PandasIndex(Index([-2303499.25, -2302499.25, -2301499.25, -2300499.25, -2299499.25,
-2298499.25, -2297499.25, -2296499.25, -2295499.25, -2294499.25,
...
2294500.75, 2295500.75, 2296500.75, 2297500.75, 2298500.75,
2299500.75, 2300500.75, 2301500.75, 2302500.75, 2303500.75],
dtype='float64', name='x', length=4608)) PandasIndex
PandasIndex(Index([-1919500.375, -1918500.375, -1917500.375, -1916500.375, -1915500.375,
-1914500.375, -1913500.375, -1912500.375, -1911500.375, -1910500.375,
...
1910499.625, 1911499.625, 1912499.625, 1913499.625, 1914499.625,
1915499.625, 1916499.625, 1917499.625, 1918499.625, 1919499.625],
dtype='float64', name='y', length=3840)) Attributes: (9)
NWM_version_number : v2.2 model_configuration : short_range model_initialization_time : 2023-04-27_11:00:00 model_output_type : forcing model_output_valid_time : 2023-04-27_12:00:00 model_total_valid_times : 18 pangeo-forge:inputs_hash : 59784c1feed1b881dc936b070ac5a83cbc680599cb9b65b9f61dae68777a174f pangeo-forge:recipe_hash : 25e9980cd34a6a0871883fc7375f224d34f403c944f9578ac5b8caea697170c8 pangeo-forge:version : 0.9.4
The actual data values at ciroh/zarr/ts/short-range-forcing-rechunked-test.zarr ciroh/short-range-forcing-kerchunk/reference.json
However, instead of a Kerchunk index file referencing the NetCDF files, this data is stored as Zarr. And notice the chunksize of 168 time
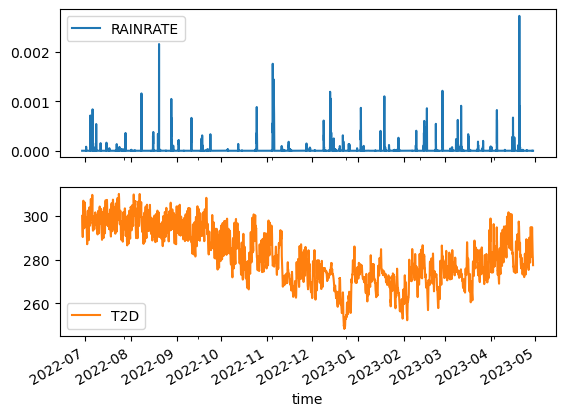
We’ll extract a timeseries for a small region of data.
NAME
STATE_NAME
geometry
0
Logan
Kentucky
POLYGON ((-87.06037 36.68085, -87.06002 36.708...
1
Queens
New York
POLYGON ((-73.96262 40.73903, -73.96243 40.739...
2
Hudson
New Jersey
MULTIPOLYGON (((-74.04220 40.69997, -74.03900 ...
3
Hunterdon
New Jersey
POLYGON ((-75.19511 40.57969, -75.19466 40.581...
4
McCreary
Kentucky
POLYGON ((-84.77845 36.60329, -84.73068 36.665...
...
...
...
...
3229
Broomfield
Colorado
MULTIPOLYGON (((-105.10667 39.95783, -105.1073...
3230
Washington
Colorado
POLYGON ((-103.70655 39.73989, -103.70655 39.7...
3231
Dakota
Minnesota
POLYGON ((-93.32967 44.77948, -93.32962 44.791...
3232
Sheridan
Nebraska
POLYGON ((-102.79287 42.82249, -102.79211 42.9...
3233
Quay
New Mexico
POLYGON ((-104.12936 34.92420, -104.12902 34.9...
3234 rows × 3 columns
<xarray.Dataset>
Dimensions: (time: 7306, x: 41, y: 40)
Coordinates:
* time (time) datetime64[ns] 2022-06-29 ... 2023-04-29T10:00:00
* x (x) float64 2.555e+05 2.565e+05 2.575e+05 ... 2.945e+05 2.955e+05
* y (y) float64 1.675e+05 1.685e+05 1.695e+05 ... 2.055e+05 2.065e+05
crs int64 0
Data variables:
LWDOWN (time, y, x) float64 dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
PSFC (time, y, x) float64 dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
Q2D (time, y, x) float64 dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
RAINRATE (time, y, x) float32 dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
SWDOWN (time, y, x) float64 dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
T2D (time, y, x) float64 dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
U2D (time, y, x) float64 dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
V2D (time, y, x) float64 dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
Attributes:
NWM_version_number: v2.2
model_configuration: short_range
model_initialization_time: 2023-04-27_11:00:00
model_output_type: forcing
model_output_valid_time: 2023-04-27_12:00:00
model_total_valid_times: 18
pangeo-forge:inputs_hash: 59784c1feed1b881dc936b070ac5a83cbc680599cb9b6...
pangeo-forge:recipe_hash: 25e9980cd34a6a0871883fc7375f224d34f403c944f95...
pangeo-forge:version: 0.9.4 Dimensions:
Coordinates: (4)
time
(time)
datetime64[ns]
2022-06-29 ... 2023-04-29T10:00:00
array(['2022-06-29T00:00:00.000000000', '2022-06-29T01:00:00.000000000',
'2022-06-29T02:00:00.000000000', ..., '2023-04-29T08:00:00.000000000',
'2023-04-29T09:00:00.000000000', '2023-04-29T10:00:00.000000000'],
dtype='datetime64[ns]') x
(x)
float64
2.555e+05 2.565e+05 ... 2.955e+05
_CoordinateAxisType : GeoX long_name : x coordinate of projection resolution : 1000.0 standard_name : projection_x_coordinate units : metre axis : X array([255500.828125, 256500.828125, 257500.828125, 258500.828125,
259500.828125, 260500.828125, 261500.828125, 262500.8125 ,
263500.8125 , 264500.8125 , 265500.8125 , 266500.8125 ,
267500.8125 , 268500.8125 , 269500.8125 , 270500.8125 ,
271500.8125 , 272500.8125 , 273500.8125 , 274500.8125 ,
275500.8125 , 276500.8125 , 277500.8125 , 278500.8125 ,
279500.8125 , 280500.8125 , 281500.8125 , 282500.8125 ,
283500.8125 , 284500.8125 , 285500.8125 , 286500.8125 ,
287500.8125 , 288500.8125 , 289500.8125 , 290500.8125 ,
291500.8125 , 292500.8125 , 293500.8125 , 294500.8125 ,
295500.8125 ]) y
(y)
float64
1.675e+05 1.685e+05 ... 2.065e+05
_CoordinateAxisType : GeoY long_name : y coordinate of projection resolution : 1000.0 standard_name : projection_y_coordinate units : metre axis : Y array([167499.65625, 168499.65625, 169499.65625, 170499.65625, 171499.65625,
172499.65625, 173499.65625, 174499.65625, 175499.65625, 176499.65625,
177499.65625, 178499.65625, 179499.65625, 180499.65625, 181499.65625,
182499.65625, 183499.65625, 184499.65625, 185499.65625, 186499.65625,
187499.65625, 188499.65625, 189499.65625, 190499.65625, 191499.65625,
192499.65625, 193499.65625, 194499.65625, 195499.65625, 196499.65625,
197499.65625, 198499.65625, 199499.65625, 200499.65625, 201499.65625,
202499.65625, 203499.65625, 204499.65625, 205499.65625, 206499.65625]) crs
()
int64
0
crs_wkt : PROJCS["Lambert_Conformal_Conic",GEOGCS["Unknown datum based upon the Authalic Sphere",DATUM["Not_specified_based_on_Authalic_Sphere",SPHEROID["Sphere",6370000,0],AUTHORITY["EPSG","6035"]],PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0],PARAMETER["false_northing",0],PARAMETER["central_meridian",-97],PARAMETER["standard_parallel_1",30],PARAMETER["standard_parallel_2",60],PARAMETER["latitude_of_origin",40],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH]] semi_major_axis : 6370000.0 semi_minor_axis : 6370000.0 inverse_flattening : 0.0 reference_ellipsoid_name : Sphere longitude_of_prime_meridian : 0.0 prime_meridian_name : Greenwich geographic_crs_name : Unknown datum based upon the Authalic Sphere horizontal_datum_name : Not specified (based on Authalic Sphere) projected_crs_name : Lambert_Conformal_Conic grid_mapping_name : lambert_conformal_conic standard_parallel : (30.0, 60.0) latitude_of_projection_origin : 40.0 longitude_of_central_meridian : -97.0 false_easting : 0.0 false_northing : 0.0 spatial_ref : PROJCS["Lambert_Conformal_Conic",GEOGCS["Unknown datum based upon the Authalic Sphere",DATUM["Not_specified_based_on_Authalic_Sphere",SPHEROID["Sphere",6370000,0],AUTHORITY["EPSG","6035"]],PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0],PARAMETER["false_northing",0],PARAMETER["central_meridian",-97],PARAMETER["standard_parallel_1",30],PARAMETER["standard_parallel_2",60],PARAMETER["latitude_of_origin",40],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH]] GeoTransform : 255000.8283203125 999.999609375 0.0 166999.65625 0.0 1000.0 Data variables: (8)
LWDOWN
(time, y, x)
float64
dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Surface downward long-wave radiation flux proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : surface_downward_longwave_flux units : W m-2
Array
Chunk
Bytes
91.41 MiB
1.69 MiB
Shape
(7306, 40, 41)
(168, 40, 33)
Dask graph
88 chunks in 6 graph layers
Data type
float64 numpy.ndarray
41
40
7306
PSFC
(time, y, x)
float64
dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Surface Pressure proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : air_pressure units : Pa
Array
Chunk
Bytes
91.41 MiB
1.69 MiB
Shape
(7306, 40, 41)
(168, 40, 33)
Dask graph
88 chunks in 6 graph layers
Data type
float64 numpy.ndarray
41
40
7306
Q2D
(time, y, x)
float64
dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : 2-m Specific Humidity proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : surface_specific_humidity units : kg kg-1
Array
Chunk
Bytes
91.41 MiB
1.69 MiB
Shape
(7306, 40, 41)
(168, 40, 33)
Dask graph
88 chunks in 6 graph layers
Data type
float64 numpy.ndarray
41
40
7306
RAINRATE
(time, y, x)
float32
dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
cell_methods : time: mean esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Surface Precipitation Rate proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : precipitation_flux units : mm s^-1
Array
Chunk
Bytes
45.71 MiB
866.25 kiB
Shape
(7306, 40, 41)
(168, 40, 33)
Dask graph
88 chunks in 6 graph layers
Data type
float32 numpy.ndarray
41
40
7306
SWDOWN
(time, y, x)
float64
dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
cell_methods : time point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : Surface downward short-wave radiation flux proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : surface_downward_shortwave_flux units : W m-2
Array
Chunk
Bytes
91.41 MiB
1.69 MiB
Shape
(7306, 40, 41)
(168, 40, 33)
Dask graph
88 chunks in 6 graph layers
Data type
float64 numpy.ndarray
41
40
7306
T2D
(time, y, x)
float64
dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : 2-m Air Temperature proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : air_temperature units : K
Array
Chunk
Bytes
91.41 MiB
1.69 MiB
Shape
(7306, 40, 41)
(168, 40, 33)
Dask graph
88 chunks in 6 graph layers
Data type
float64 numpy.ndarray
41
40
7306
U2D
(time, y, x)
float64
dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : 10-m U-component of wind proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : x_wind units : m s-1
Array
Chunk
Bytes
91.41 MiB
1.69 MiB
Shape
(7306, 40, 41)
(168, 40, 33)
Dask graph
88 chunks in 6 graph layers
Data type
float64 numpy.ndarray
41
40
7306
V2D
(time, y, x)
float64
dask.array<chunksize=(168, 40, 33), meta=np.ndarray>
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : 10-m V-component of wind proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : y_wind units : m s-1
Array
Chunk
Bytes
91.41 MiB
1.69 MiB
Shape
(7306, 40, 41)
(168, 40, 33)
Dask graph
88 chunks in 6 graph layers
Data type
float64 numpy.ndarray
41
40
7306
Indexes: (3)
PandasIndex
PandasIndex(DatetimeIndex(['2022-06-29 00:00:00', '2022-06-29 01:00:00',
'2022-06-29 02:00:00', '2022-06-29 03:00:00',
'2022-06-29 04:00:00', '2022-06-29 05:00:00',
'2022-06-29 06:00:00', '2022-06-29 07:00:00',
'2022-06-29 08:00:00', '2022-06-29 09:00:00',
...
'2023-04-29 01:00:00', '2023-04-29 02:00:00',
'2023-04-29 03:00:00', '2023-04-29 04:00:00',
'2023-04-29 05:00:00', '2023-04-29 06:00:00',
'2023-04-29 07:00:00', '2023-04-29 08:00:00',
'2023-04-29 09:00:00', '2023-04-29 10:00:00'],
dtype='datetime64[ns]', name='time', length=7306, freq=None)) PandasIndex
PandasIndex(Index([255500.828125, 256500.828125, 257500.828125, 258500.828125,
259500.828125, 260500.828125, 261500.828125, 262500.8125,
263500.8125, 264500.8125, 265500.8125, 266500.8125,
267500.8125, 268500.8125, 269500.8125, 270500.8125,
271500.8125, 272500.8125, 273500.8125, 274500.8125,
275500.8125, 276500.8125, 277500.8125, 278500.8125,
279500.8125, 280500.8125, 281500.8125, 282500.8125,
283500.8125, 284500.8125, 285500.8125, 286500.8125,
287500.8125, 288500.8125, 289500.8125, 290500.8125,
291500.8125, 292500.8125, 293500.8125, 294500.8125,
295500.8125],
dtype='float64', name='x')) PandasIndex
PandasIndex(Index([167499.65625, 168499.65625, 169499.65625, 170499.65625, 171499.65625,
172499.65625, 173499.65625, 174499.65625, 175499.65625, 176499.65625,
177499.65625, 178499.65625, 179499.65625, 180499.65625, 181499.65625,
182499.65625, 183499.65625, 184499.65625, 185499.65625, 186499.65625,
187499.65625, 188499.65625, 189499.65625, 190499.65625, 191499.65625,
192499.65625, 193499.65625, 194499.65625, 195499.65625, 196499.65625,
197499.65625, 198499.65625, 199499.65625, 200499.65625, 201499.65625,
202499.65625, 203499.65625, 204499.65625, 205499.65625, 206499.65625],
dtype='float64', name='y')) Attributes: (9)
NWM_version_number : v2.2 model_configuration : short_range model_initialization_time : 2023-04-27_11:00:00 model_output_type : forcing model_output_valid_time : 2023-04-27_12:00:00 model_total_valid_times : 18 pangeo-forge:inputs_hash : 59784c1feed1b881dc936b070ac5a83cbc680599cb9b65b9f61dae68777a174f pangeo-forge:recipe_hash : 25e9980cd34a6a0871883fc7375f224d34f403c944f9578ac5b8caea697170c8 pangeo-forge:version : 0.9.4
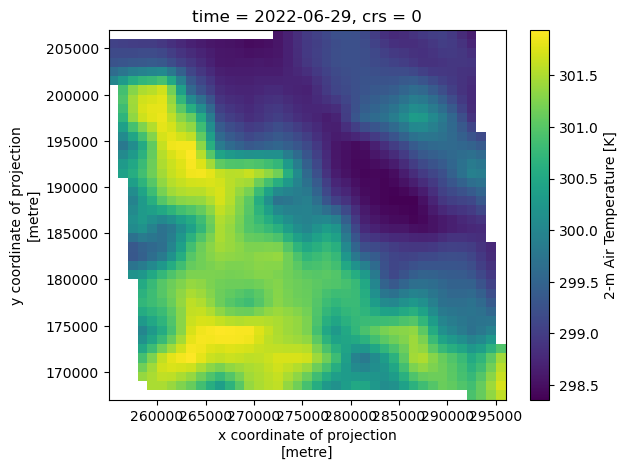
Accessing a single timestamp of data from this subset is fast:
Accessing data through time is also relatively fast (compared to using the dataset that’s chunked in time).
Client
Client-632558ef-f4bf-11ed-83c7-32bb6486e0f8
Launch dashboard in JupyterLab
Cluster Info
LocalCluster
67d77040
Scheduler Info
Scheduler
Scheduler-f6320a91-76fc-4263-add6-d560952fadca
Workers
Worker: 0
Comm: tcp://127.0.0.1:46111
Total threads: 2
Dashboard: /user/taugspurger/proxy/34917/status
Memory: 1.88 GiB
Nanny: tcp://127.0.0.1:33065
Local directory: /tmp/dask-worker-space/worker-vumqckxz
Worker: 1
Comm: tcp://127.0.0.1:45429
Total threads: 2
Dashboard: /user/taugspurger/proxy/44995/status
Memory: 1.88 GiB
Nanny: tcp://127.0.0.1:34179
Local directory: /tmp/dask-worker-space/worker-xbn6qzu4
Worker: 2
Comm: tcp://127.0.0.1:38817
Total threads: 2
Dashboard: /user/taugspurger/proxy/41733/status
Memory: 1.88 GiB
Nanny: tcp://127.0.0.1:39513
Local directory: /tmp/dask-worker-space/worker-_k5g9cyp
Worker: 3
Comm: tcp://127.0.0.1:35347
Total threads: 2
Dashboard: /user/taugspurger/proxy/40943/status
Memory: 1.88 GiB
Nanny: tcp://127.0.0.1:45679
Local directory: /tmp/dask-worker-space/worker-ob5iaaus
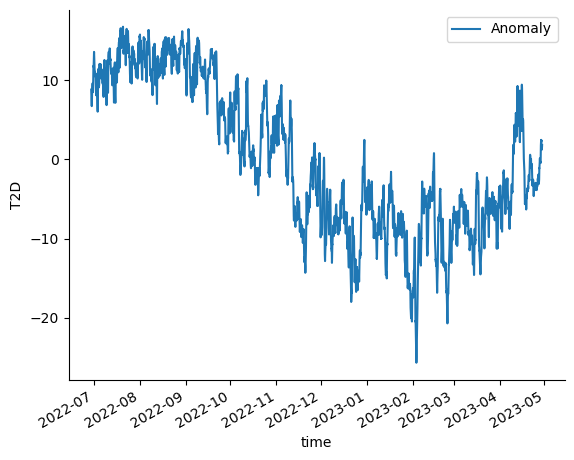
Now let’s do a fun computation. We’ll calculate an average temperature at each hour, and look at the difference (anomaly?) from it over the course of our timeseries.
This will be a large computation, so let’s parallelize it on a cluster.
Client
Client-7e49bf19-f4bf-11ed-83c7-32bb6486e0f8
Launch dashboard in JupyterLab
Cluster Info
<xarray.DataArray 'T2D' (time: 7306, y: 480, x: 576)>
dask.array<getitem, shape=(7306, 480, 576), dtype=float64, chunksize=(168, 240, 288), chunktype=numpy.ndarray>
Coordinates:
crs int64 0
* time (time) datetime64[ns] 2022-06-29 ... 2023-04-29T10:00:00
* x (x) float64 5.765e+05 5.775e+05 5.785e+05 ... 1.151e+06 1.152e+06
* y (y) float64 4.805e+05 4.815e+05 4.825e+05 ... 9.585e+05 9.595e+05
Attributes:
cell_methods: time: point
esri_pe_string: PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DAT...
grid_mapping: crs
long_name: 2-m Air Temperature
proj4: +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0...
remap: remapped via ESMF regrid_with_weights: Bilinear
standard_name: air_temperature
units: K Coordinates: (4)
crs
()
int64
0
crs_wkt : PROJCS["Lambert_Conformal_Conic",GEOGCS["Unknown datum based upon the Authalic Sphere",DATUM["Not_specified_based_on_Authalic_Sphere",SPHEROID["Sphere",6370000,0],AUTHORITY["EPSG","6035"]],PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0],PARAMETER["false_northing",0],PARAMETER["central_meridian",-97],PARAMETER["standard_parallel_1",30],PARAMETER["standard_parallel_2",60],PARAMETER["latitude_of_origin",40],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH]] semi_major_axis : 6370000.0 semi_minor_axis : 6370000.0 inverse_flattening : 0.0 reference_ellipsoid_name : Sphere longitude_of_prime_meridian : 0.0 prime_meridian_name : Greenwich geographic_crs_name : Unknown datum based upon the Authalic Sphere horizontal_datum_name : Not specified (based on Authalic Sphere) projected_crs_name : Lambert_Conformal_Conic grid_mapping_name : lambert_conformal_conic standard_parallel : (30.0, 60.0) latitude_of_projection_origin : 40.0 longitude_of_central_meridian : -97.0 false_easting : 0.0 false_northing : 0.0 spatial_ref : PROJCS["Lambert_Conformal_Conic",GEOGCS["Unknown datum based upon the Authalic Sphere",DATUM["Not_specified_based_on_Authalic_Sphere",SPHEROID["Sphere",6370000,0],AUTHORITY["EPSG","6035"]],PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0],PARAMETER["false_northing",0],PARAMETER["central_meridian",-97],PARAMETER["standard_parallel_1",30],PARAMETER["standard_parallel_2",60],PARAMETER["latitude_of_origin",40],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH]] time
(time)
datetime64[ns]
2022-06-29 ... 2023-04-29T10:00:00
array(['2022-06-29T00:00:00.000000000', '2022-06-29T01:00:00.000000000',
'2022-06-29T02:00:00.000000000', ..., '2023-04-29T08:00:00.000000000',
'2023-04-29T09:00:00.000000000', '2023-04-29T10:00:00.000000000'],
dtype='datetime64[ns]') x
(x)
float64
5.765e+05 5.775e+05 ... 1.152e+06
_CoordinateAxisType : GeoX long_name : x coordinate of projection resolution : 1000.0 standard_name : projection_x_coordinate units : m array([ 576500.8125, 577500.8125, 578500.8125, ..., 1149500.875 ,
1150500.875 , 1151500.875 ]) y
(y)
float64
4.805e+05 4.815e+05 ... 9.595e+05
_CoordinateAxisType : GeoY long_name : y coordinate of projection resolution : 1000.0 standard_name : projection_y_coordinate units : m array([480499.65625, 481499.65625, 482499.65625, ..., 957499.6875 ,
958499.6875 , 959499.6875 ]) Indexes: (3)
PandasIndex
PandasIndex(DatetimeIndex(['2022-06-29 00:00:00', '2022-06-29 01:00:00',
'2022-06-29 02:00:00', '2022-06-29 03:00:00',
'2022-06-29 04:00:00', '2022-06-29 05:00:00',
'2022-06-29 06:00:00', '2022-06-29 07:00:00',
'2022-06-29 08:00:00', '2022-06-29 09:00:00',
...
'2023-04-29 01:00:00', '2023-04-29 02:00:00',
'2023-04-29 03:00:00', '2023-04-29 04:00:00',
'2023-04-29 05:00:00', '2023-04-29 06:00:00',
'2023-04-29 07:00:00', '2023-04-29 08:00:00',
'2023-04-29 09:00:00', '2023-04-29 10:00:00'],
dtype='datetime64[ns]', name='time', length=7306, freq=None)) PandasIndex
PandasIndex(Index([576500.8125, 577500.8125, 578500.8125, 579500.8125, 580500.8125,
581500.8125, 582500.8125, 583500.8125, 584500.8125, 585500.8125,
...
1142500.875, 1143500.875, 1144500.875, 1145500.875, 1146500.875,
1147500.875, 1148500.875, 1149500.875, 1150500.875, 1151500.875],
dtype='float64', name='x', length=576)) PandasIndex
PandasIndex(Index([480499.65625, 481499.65625, 482499.65625, 483499.65625, 484499.65625,
485499.65625, 486499.65625, 487499.65625, 488499.65625, 489499.65625,
...
950499.6875, 951499.6875, 952499.6875, 953499.6875, 954499.6875,
955499.6875, 956499.6875, 957499.6875, 958499.6875, 959499.6875],
dtype='float64', name='y', length=480)) Attributes: (8)
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : 2-m Air Temperature proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : air_temperature units : K
<xarray.DataArray 'T2D' (hour: 24, y: 480, x: 576)>
dask.array<transpose, shape=(24, 480, 576), dtype=float64, chunksize=(24, 240, 288), chunktype=numpy.ndarray>
Coordinates:
crs int64 0
* x (x) float64 5.765e+05 5.775e+05 5.785e+05 ... 1.151e+06 1.152e+06
* y (y) float64 4.805e+05 4.815e+05 4.825e+05 ... 9.585e+05 9.595e+05
* hour (hour) int64 0 1 2 3 4 5 6 7 8 9 ... 14 15 16 17 18 19 20 21 22 23
Attributes:
cell_methods: time: point
esri_pe_string: PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DAT...
grid_mapping: crs
long_name: 2-m Air Temperature
proj4: +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0...
remap: remapped via ESMF regrid_with_weights: Bilinear
standard_name: air_temperature
units: K Coordinates: (4)
crs
()
int64
0
crs_wkt : PROJCS["Lambert_Conformal_Conic",GEOGCS["Unknown datum based upon the Authalic Sphere",DATUM["Not_specified_based_on_Authalic_Sphere",SPHEROID["Sphere",6370000,0],AUTHORITY["EPSG","6035"]],PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0],PARAMETER["false_northing",0],PARAMETER["central_meridian",-97],PARAMETER["standard_parallel_1",30],PARAMETER["standard_parallel_2",60],PARAMETER["latitude_of_origin",40],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH]] semi_major_axis : 6370000.0 semi_minor_axis : 6370000.0 inverse_flattening : 0.0 reference_ellipsoid_name : Sphere longitude_of_prime_meridian : 0.0 prime_meridian_name : Greenwich geographic_crs_name : Unknown datum based upon the Authalic Sphere horizontal_datum_name : Not specified (based on Authalic Sphere) projected_crs_name : Lambert_Conformal_Conic grid_mapping_name : lambert_conformal_conic standard_parallel : (30.0, 60.0) latitude_of_projection_origin : 40.0 longitude_of_central_meridian : -97.0 false_easting : 0.0 false_northing : 0.0 spatial_ref : PROJCS["Lambert_Conformal_Conic",GEOGCS["Unknown datum based upon the Authalic Sphere",DATUM["Not_specified_based_on_Authalic_Sphere",SPHEROID["Sphere",6370000,0],AUTHORITY["EPSG","6035"]],PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0],PARAMETER["false_northing",0],PARAMETER["central_meridian",-97],PARAMETER["standard_parallel_1",30],PARAMETER["standard_parallel_2",60],PARAMETER["latitude_of_origin",40],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH]] x
(x)
float64
5.765e+05 5.775e+05 ... 1.152e+06
_CoordinateAxisType : GeoX long_name : x coordinate of projection resolution : 1000.0 standard_name : projection_x_coordinate units : m array([ 576500.8125, 577500.8125, 578500.8125, ..., 1149500.875 ,
1150500.875 , 1151500.875 ]) y
(y)
float64
4.805e+05 4.815e+05 ... 9.595e+05
_CoordinateAxisType : GeoY long_name : y coordinate of projection resolution : 1000.0 standard_name : projection_y_coordinate units : m array([480499.65625, 481499.65625, 482499.65625, ..., 957499.6875 ,
958499.6875 , 959499.6875 ]) hour
(hour)
int64
0 1 2 3 4 5 6 ... 18 19 20 21 22 23
array([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23]) Indexes: (3)
PandasIndex
PandasIndex(Index([576500.8125, 577500.8125, 578500.8125, 579500.8125, 580500.8125,
581500.8125, 582500.8125, 583500.8125, 584500.8125, 585500.8125,
...
1142500.875, 1143500.875, 1144500.875, 1145500.875, 1146500.875,
1147500.875, 1148500.875, 1149500.875, 1150500.875, 1151500.875],
dtype='float64', name='x', length=576)) PandasIndex
PandasIndex(Index([480499.65625, 481499.65625, 482499.65625, 483499.65625, 484499.65625,
485499.65625, 486499.65625, 487499.65625, 488499.65625, 489499.65625,
...
950499.6875, 951499.6875, 952499.6875, 953499.6875, 954499.6875,
955499.6875, 956499.6875, 957499.6875, 958499.6875, 959499.6875],
dtype='float64', name='y', length=480)) PandasIndex
PandasIndex(Index([ 0, 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17,
18, 19, 20, 21, 22, 23],
dtype='int64', name='hour')) Attributes: (8)
cell_methods : time: point esri_pe_string : PROJCS["Lambert_Conformal_Conic",GEOGCS["GCS_Sphere",DATUM["D_Sphere",SPHEROID["Sphere",6370000.0,0.0]],PRIMEM["Greenwich",0.0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0.0],PARAMETER["false_northing",0.0],PARAMETER["central_meridian",-97.0],PARAMETER["standard_parallel_1",30.0],PARAMETER["standard_parallel_2",60.0],PARAMETER["latitude_of_origin",40.0],UNIT["Meter",1.0]];-35691800 -29075200 10000;-100000 10000;-100000 10000;0.001;0.001;0.001;IsHighPrecision grid_mapping : crs long_name : 2-m Air Temperature proj4 : +proj=lcc +units=m +a=6370000.0 +b=6370000.0 +lat_1=30.0 +lat_2=60.0 +lat_0=40.0 +lon_0=-97.0 +x_0=0 +y_0=0 +k_0=1.0 +nadgrids=@null +wktext +no_defs remap : remapped via ESMF regrid_with_weights: Bilinear standard_name : air_temperature units : K
We’ll use this result in a computation later on, so let’s persist it on the cluster.
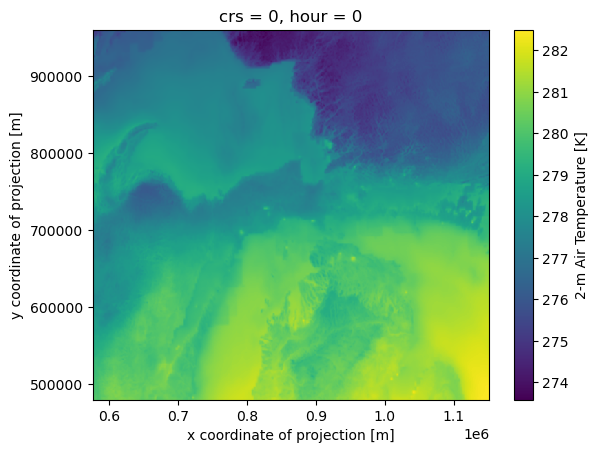
And peek at the first timestamp.
Now we can calculate the anomaly.
<xarray.DataArray 'T2D' (time: 7306, y: 480, x: 576)>
dask.array<sub, shape=(7306, 480, 576), dtype=float64, chunksize=(168, 240, 288), chunktype=numpy.ndarray>
Coordinates:
crs (time) int64 0 0 0 0 0 0 0 0 0 0 0 0 0 ... 0 0 0 0 0 0 0 0 0 0 0 0
* time (time) datetime64[ns] 2022-06-29 ... 2023-04-29T10:00:00
* x (x) float64 5.765e+05 5.775e+05 5.785e+05 ... 1.151e+06 1.152e+06
* y (y) float64 4.805e+05 4.815e+05 4.825e+05 ... 9.585e+05 9.595e+05
hour (time) int64 0 1 2 3 4 5 6 7 8 9 10 11 ... 0 1 2 3 4 5 6 7 8 9 10 Coordinates: (5)
crs
(time)
int64
0 0 0 0 0 0 0 0 ... 0 0 0 0 0 0 0 0
crs_wkt : PROJCS["Lambert_Conformal_Conic",GEOGCS["Unknown datum based upon the Authalic Sphere",DATUM["Not_specified_based_on_Authalic_Sphere",SPHEROID["Sphere",6370000,0],AUTHORITY["EPSG","6035"]],PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0],PARAMETER["false_northing",0],PARAMETER["central_meridian",-97],PARAMETER["standard_parallel_1",30],PARAMETER["standard_parallel_2",60],PARAMETER["latitude_of_origin",40],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH]] semi_major_axis : 6370000.0 semi_minor_axis : 6370000.0 inverse_flattening : 0.0 reference_ellipsoid_name : Sphere longitude_of_prime_meridian : 0.0 prime_meridian_name : Greenwich geographic_crs_name : Unknown datum based upon the Authalic Sphere horizontal_datum_name : Not specified (based on Authalic Sphere) projected_crs_name : Lambert_Conformal_Conic grid_mapping_name : lambert_conformal_conic standard_parallel : (30.0, 60.0) latitude_of_projection_origin : 40.0 longitude_of_central_meridian : -97.0 false_easting : 0.0 false_northing : 0.0 spatial_ref : PROJCS["Lambert_Conformal_Conic",GEOGCS["Unknown datum based upon the Authalic Sphere",DATUM["Not_specified_based_on_Authalic_Sphere",SPHEROID["Sphere",6370000,0],AUTHORITY["EPSG","6035"]],PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0],PARAMETER["false_northing",0],PARAMETER["central_meridian",-97],PARAMETER["standard_parallel_1",30],PARAMETER["standard_parallel_2",60],PARAMETER["latitude_of_origin",40],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH]] array([0, 0, 0, ..., 0, 0, 0]) time
(time)
datetime64[ns]
2022-06-29 ... 2023-04-29T10:00:00
array(['2022-06-29T00:00:00.000000000', '2022-06-29T01:00:00.000000000',
'2022-06-29T02:00:00.000000000', ..., '2023-04-29T08:00:00.000000000',
'2023-04-29T09:00:00.000000000', '2023-04-29T10:00:00.000000000'],
dtype='datetime64[ns]') x
(x)
float64
5.765e+05 5.775e+05 ... 1.152e+06
_CoordinateAxisType : GeoX long_name : x coordinate of projection resolution : 1000.0 standard_name : projection_x_coordinate units : m array([ 576500.8125, 577500.8125, 578500.8125, ..., 1149500.875 ,
1150500.875 , 1151500.875 ]) y
(y)
float64
4.805e+05 4.815e+05 ... 9.595e+05
_CoordinateAxisType : GeoY long_name : y coordinate of projection resolution : 1000.0 standard_name : projection_y_coordinate units : m array([480499.65625, 481499.65625, 482499.65625, ..., 957499.6875 ,
958499.6875 , 959499.6875 ]) hour
(time)
int64
0 1 2 3 4 5 6 7 ... 4 5 6 7 8 9 10
array([ 0, 1, 2, ..., 8, 9, 10]) Indexes: (3)
PandasIndex
PandasIndex(DatetimeIndex(['2022-06-29 00:00:00', '2022-06-29 01:00:00',
'2022-06-29 02:00:00', '2022-06-29 03:00:00',
'2022-06-29 04:00:00', '2022-06-29 05:00:00',
'2022-06-29 06:00:00', '2022-06-29 07:00:00',
'2022-06-29 08:00:00', '2022-06-29 09:00:00',
...
'2023-04-29 01:00:00', '2023-04-29 02:00:00',
'2023-04-29 03:00:00', '2023-04-29 04:00:00',
'2023-04-29 05:00:00', '2023-04-29 06:00:00',
'2023-04-29 07:00:00', '2023-04-29 08:00:00',
'2023-04-29 09:00:00', '2023-04-29 10:00:00'],
dtype='datetime64[ns]', name='time', length=7306, freq=None)) PandasIndex
PandasIndex(Index([576500.8125, 577500.8125, 578500.8125, 579500.8125, 580500.8125,
581500.8125, 582500.8125, 583500.8125, 584500.8125, 585500.8125,
...
1142500.875, 1143500.875, 1144500.875, 1145500.875, 1146500.875,
1147500.875, 1148500.875, 1149500.875, 1150500.875, 1151500.875],
dtype='float64', name='x', length=576)) PandasIndex
PandasIndex(Index([480499.65625, 481499.65625, 482499.65625, 483499.65625, 484499.65625,
485499.65625, 486499.65625, 487499.65625, 488499.65625, 489499.65625,
...
950499.6875, 951499.6875, 952499.6875, 953499.6875, 954499.6875,
955499.6875, 956499.6875, 957499.6875, 958499.6875, 959499.6875],
dtype='float64', name='y', length=480)) Attributes: (0)
<xarray.DataArray 'T2D' (time: 7306)>
dask.array<mean_agg-aggregate, shape=(7306,), dtype=float64, chunksize=(168,), chunktype=numpy.ndarray>
Coordinates:
crs (time) int64 0 0 0 0 0 0 0 0 0 0 0 0 0 ... 0 0 0 0 0 0 0 0 0 0 0 0
* time (time) datetime64[ns] 2022-06-29 ... 2023-04-29T10:00:00
hour (time) int64 0 1 2 3 4 5 6 7 8 9 10 11 ... 0 1 2 3 4 5 6 7 8 9 10 Coordinates: (3)
crs
(time)
int64
0 0 0 0 0 0 0 0 ... 0 0 0 0 0 0 0 0
crs_wkt : PROJCS["Lambert_Conformal_Conic",GEOGCS["Unknown datum based upon the Authalic Sphere",DATUM["Not_specified_based_on_Authalic_Sphere",SPHEROID["Sphere",6370000,0],AUTHORITY["EPSG","6035"]],PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0],PARAMETER["false_northing",0],PARAMETER["central_meridian",-97],PARAMETER["standard_parallel_1",30],PARAMETER["standard_parallel_2",60],PARAMETER["latitude_of_origin",40],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH]] semi_major_axis : 6370000.0 semi_minor_axis : 6370000.0 inverse_flattening : 0.0 reference_ellipsoid_name : Sphere longitude_of_prime_meridian : 0.0 prime_meridian_name : Greenwich geographic_crs_name : Unknown datum based upon the Authalic Sphere horizontal_datum_name : Not specified (based on Authalic Sphere) projected_crs_name : Lambert_Conformal_Conic grid_mapping_name : lambert_conformal_conic standard_parallel : (30.0, 60.0) latitude_of_projection_origin : 40.0 longitude_of_central_meridian : -97.0 false_easting : 0.0 false_northing : 0.0 spatial_ref : PROJCS["Lambert_Conformal_Conic",GEOGCS["Unknown datum based upon the Authalic Sphere",DATUM["Not_specified_based_on_Authalic_Sphere",SPHEROID["Sphere",6370000,0],AUTHORITY["EPSG","6035"]],PRIMEM["Greenwich",0],UNIT["Degree",0.0174532925199433]],PROJECTION["Lambert_Conformal_Conic_2SP"],PARAMETER["false_easting",0],PARAMETER["false_northing",0],PARAMETER["central_meridian",-97],PARAMETER["standard_parallel_1",30],PARAMETER["standard_parallel_2",60],PARAMETER["latitude_of_origin",40],UNIT["metre",1,AUTHORITY["EPSG","9001"]],AXIS["Easting",EAST],AXIS["Northing",NORTH]] array([0, 0, 0, ..., 0, 0, 0]) time
(time)
datetime64[ns]
2022-06-29 ... 2023-04-29T10:00:00
array(['2022-06-29T00:00:00.000000000', '2022-06-29T01:00:00.000000000',
'2022-06-29T02:00:00.000000000', ..., '2023-04-29T08:00:00.000000000',
'2023-04-29T09:00:00.000000000', '2023-04-29T10:00:00.000000000'],
dtype='datetime64[ns]') hour
(time)
int64
0 1 2 3 4 5 6 7 ... 4 5 6 7 8 9 10
array([ 0, 1, 2, ..., 8, 9, 10]) Indexes: (1)
PandasIndex
PandasIndex(DatetimeIndex(['2022-06-29 00:00:00', '2022-06-29 01:00:00',
'2022-06-29 02:00:00', '2022-06-29 03:00:00',
'2022-06-29 04:00:00', '2022-06-29 05:00:00',
'2022-06-29 06:00:00', '2022-06-29 07:00:00',
'2022-06-29 08:00:00', '2022-06-29 09:00:00',
...
'2023-04-29 01:00:00', '2023-04-29 02:00:00',
'2023-04-29 03:00:00', '2023-04-29 04:00:00',
'2023-04-29 05:00:00', '2023-04-29 06:00:00',
'2023-04-29 07:00:00', '2023-04-29 08:00:00',
'2023-04-29 09:00:00', '2023-04-29 10:00:00'],
dtype='datetime64[ns]', name='time', length=7306, freq=None)) Attributes: (0)
This is a small result, so we can bring it back to the client with .compute()
And visualize the result.
Now let’s clean up and move on to Tabular